
And nowhere has that need been more evident than in the field of next-generation sequencing. We can provide gene decoding and analysis for whole genome, exome. More data create more opportunities and a more pressing need for systematic methods of analysis. Office hours will be held once between sessions to help with homework questions or general questions about the previous session. Our NGS Service provides wide range of NGS applications for various research purposes. PlasmidID is a mapping-based, assembly-assisted plasmid identification tool that analyzes and gives graphic solution for plasmid identification. Homework assignments for practicing key concepts will be assigned at the end of each session, and they will be due at the beginning of the following session in one week (homework assignments will take about 4 hours to complete).
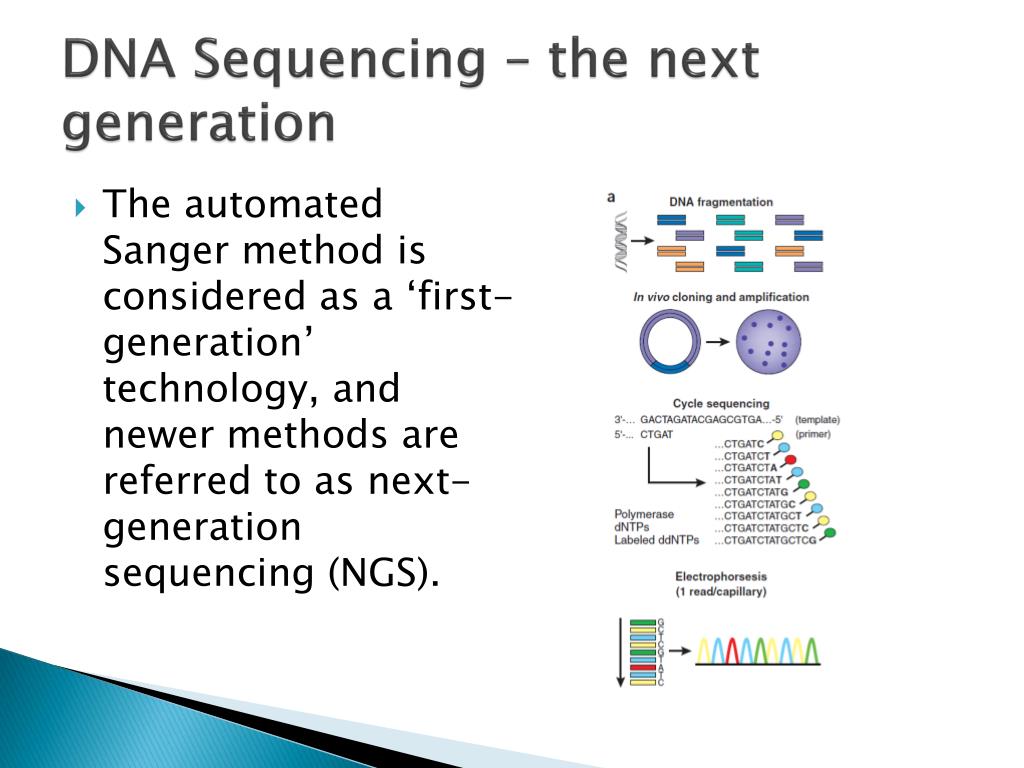
The different NGS platforms, its applications and different sequencing. Please note that attendance is mandatory for the first 5 sessions (see schedule below for dates). In this paper work flow of data analysis part of NGS are clearly discussed. Each session runs for 2 consecutive days (9am to 5pm), and the sessions are held every week. The course is comprised of five hands-on sessions held over a period of five weeks beginning mid-June (2016). Prior programming experience or command-line training is not required to take this course. During this course, participants will gain skills in the areas of (a) UNIX and basic shell scripting, (b) high-performance compute clusters, and (c) R for statistical analysis and data visualization. At the end of this course, participants can expect to have the expertise to independently run data analysis for sequencing experiments. This self-paced, introductory course, aimed at professionals who want to break into the sequencing-related field of bioinformatics, explores important public. The critical difference is that NGS sequences millions of fragments in a massively parallel fashion, improving speed and accuracy while reducing the cost of sequencing. In principle, the concept is similar to capillary electrophoresis. The sessions will also include functional analysis downstream of sequence data processing. The basic next-generation sequencing process involves fragmenting DNA/RNA into multiple pieces, adding adapters, sequencing the libraries, and reassembling them to form a genomic sequence. The topics covered in-depth during this course are analysis of RNA-Seq and ChIP-Seq data, with an optional Variant Calling session. Questions? Contact course is aimed at bench biologists who are interested in learning about NGS-based genomic analysis. This is open to members of HSCI, HMS affiliated researchers from the Basic and Social Science Depts. Visit the course webpage for complete details. The pre-registration deadline for this course is Sunday, May 1, 2016.
Program of the Harvard Chan Bioinformatics Core


 0 kommentar(er)
0 kommentar(er)
